Gaussian Naive Bayes (GNB) is a classification technique used in machine learning based on a probabilistic approach and Gaussian distribution. Gaussian Naive Bayes assumes that each parameter, also called features or predictors, has an independent capacity of predicting the output variable.
What Is Gaussian Naive Bayes Classifier?
Gaussian Naive Bayes is a machine learning classification technique based on a probabilistic approach that assumes each class follows a normal distribution. It assumes each parameter has an independent capacity of predicting the output variable. It is able to predict the probability of a dependent variable to be classified in each group.
The combination of the prediction for all parameters is the final prediction that returns a probability of the dependent variable to be classified in each group. The final classification is assigned to the group with the higher probability.
What Are Naive Bayes Classifiers?
Naive Bayes classifiers are supervised machine learning algorithms used to solve classification problems. The term “naive” comes from the fact that Naive Bayes classifiers assume all features in a model are independent, meaning they do not affect one another. They also assume each feature plays an equal role in contributing to a model’s output, so they cannot discern the most important features in a model.
These classifiers contain a small number of parameters, making them simpler and faster than other types of classifiers. As a result, Naive Bayes classifiers are easy to work with and great to introduce to machine learning beginners. While there are wide variations of Naive Bayes classifiers, one of the most popular types is the Gaussian Naive Bayes classifier.
What Is Gaussian Distribution?
Gaussian distribution is also called normal distribution. Normal distribution is a statistical model that describes the distributions of continuous random variables in nature and is defined by its bell-shaped curve. The two most important features of the normal distribution are the mean (μ) and standard deviation (σ). The mean is the average value of a distribution, and the standard deviation is the “width” of the distribution around the mean.
A variable (X) that is normally distributed is distributed continuously (continuous variable) from −∞ < X < +∞, and the total area under the model curve is 1.
How to Use Gaussian Naive Bayes for Multi-Classification in Scikit-Learn
Gaussian Naive Bayes classification algorithm requires just a few steps to complete for multi-classification. Here’s how to do it yourself with sample code.
1. Import the Libraries
The first step is to import the necessary libraries:
from random import random
from random import randint
import pandas as pd
import numpy as np
import seaborn as sns
import matplotlib.pyplot as plt
import statistics
from sklearn.model_selection import train_test_split
from sklearn.preprocessing import StandardScaler
from sklearn.naive_bayes import GaussianNB
from sklearn.metrics import confusion_matrix
from mlxtend.plotting import plot_decision_regions
2. Create a Database
Now, we will create a database in which the predictive variables are normally distributed.
#Creating values for FeNO with 3 classes:
FeNO_0 = np.random.normal(20, 19, 200)
FeNO_1 = np.random.normal(40, 20, 200)
FeNO_2 = np.random.normal(60, 20, 200)
#Creating values for FEV1 with 3 classes:
FEV1_0 = np.random.normal(4.65, 1, 200)
FEV1_1 = np.random.normal(3.75, 1.2, 200)
FEV1_2 = np.random.normal(2.85, 1.2, 200)
#Creating values for Broncho Dilation with 3 classes:
BD_0 = np.random.normal(150,49, 200)
BD_1 = np.random.normal(201,50, 200)
BD_2 = np.random.normal(251, 50, 200)
#Creating labels variable with three classes:(2)disease (1)possible disease (0)no disease:
not_asthma = np.zeros((200,), dtype=int)
poss_asthma = np.ones((200,), dtype=int)
asthma = np.full((200,), 2, dtype=int)
#Concatenate classes into one variable:
FeNO = np.concatenate([FeNO_0, FeNO_1, FeNO_2])
FEV1 = np.concatenate([FEV1_0, FEV1_1, FEV1_2])
BD = np.concatenate([BD_0, BD_1, BD_2])
dx = np.concatenate([not_asthma, poss_asthma, asthma])
#Create DataFrame:
df = pd.DataFrame()
#Add variables to DataFrame:
df['FeNO'] = FeNO.tolist()
df['FEV1'] = FEV1.tolist()
df['BD'] = BD.tolist()
df['dx'] = dx.tolist()
#Check database:
df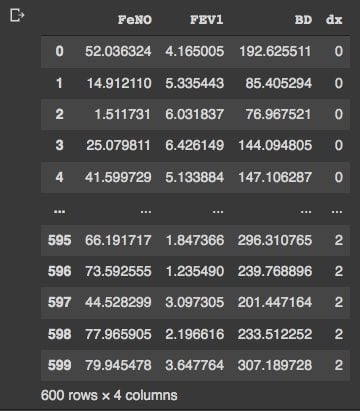
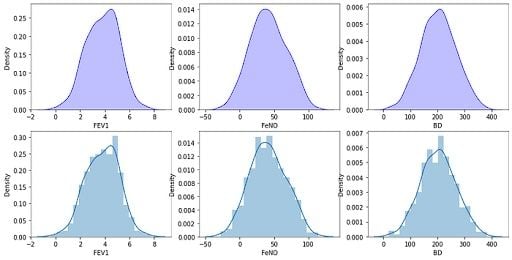
3. Check the Distribution of Variables
We have our data frame with 600 rows and four columns. Now, we can check the distribution of our variables by visual inspection:
fig, axs = plt.subplots(2, 3, figsize=(14, 7))
sns.kdeplot(df['FEV1'], shade=True, color="b", ax=axs[0, 0])
sns.kdeplot(df['FeNO'], shade=True, color="b", ax=axs[0, 1])
sns.kdeplot(df['BD'], shade=True, color="b", ax=axs[0, 2])
sns.distplot( a=df["FEV1"], hist=True, kde=True, rug=False, ax=axs[1, 0])
sns.distplot( a=df["FeNO"], hist=True, kde=True, rug=False, ax=axs[1, 1])
sns.distplot( a=df["BD"], hist=True, kde=True, rug=False, ax=axs[1, 2])
plt.show()
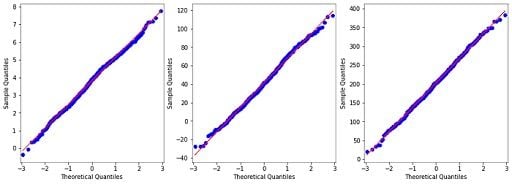
4. Build QQ-Plots to Double-Check Gaussian Distribution
By visual inspection, our data seems to be close to the Gaussian distribution. We can double-check building qq-plots:
from statsmodels.graphics.gofplots import qqplot
from matplotlib import pyplot
#q-q plot:
fig, axs = pyplot.subplots(1, 3, figsize=(15, 5))
qqplot(df['FEV1'], line='s', ax=axs[0])
qqplot(df['FeNO'], line='s', ax=axs[1])
qqplot(df['BD'], line='s', ax=axs[2])
pyplot.show()
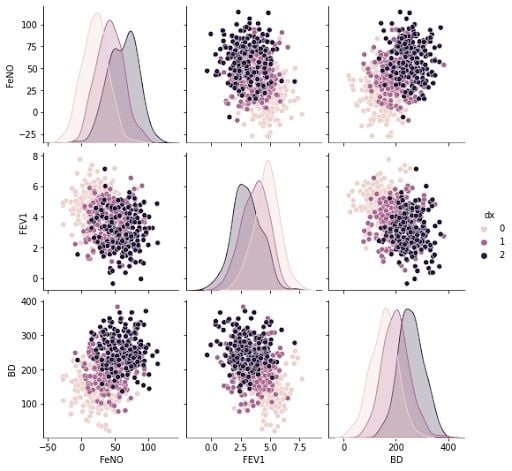
5. Build a Pair-Plot to Explore the Data Set
We don’t have a perfect normal distribution for our variables, but we are close enough and able to use them. To explore our data set and the correlations between variables, we will build a pair-plot:
#Exploring dataset:
sns.pairplot(df, kind="scatter", hue="dx")
plt.show()
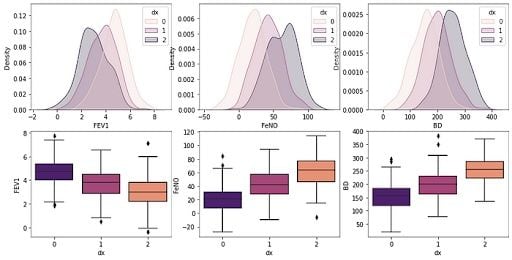
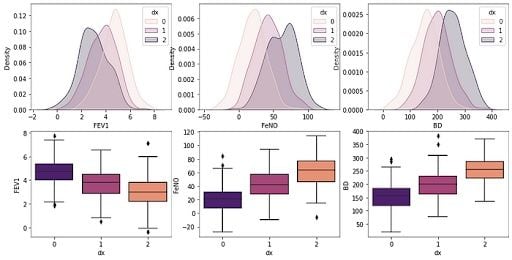
6. Create a Box-Plot to Check Parameters
Next we’ll build a boxplot graph to check the distributions for the three groups and see which parameters best distinguish the categories:
# plotting both distibutions on the same figure
fig, axs = plt.subplots(2, 3, figsize=(14, 7))
fig = sns.kdeplot(df['FEV1'], hue= df['dx'], shade=True, color="r", ax=axs[0, 0])
fig = sns.kdeplot(df['FeNO'], hue= df['dx'], shade=True, color="r", ax=axs[0, 1])
fig = sns.kdeplot(df['BD'], hue= df['dx'], shade=True, color="r", ax=axs[0, 2])
sns.boxplot(x=df["dx"], y=df["FEV1"], palette = 'magma', ax=axs[1, 0])
sns.boxplot(x=df["dx"], y=df["FeNO"], palette = 'magma',ax=axs[1, 1])
sns.boxplot(x=df["dx"], y=df["BD"], palette = 'magma',ax=axs[1, 2])
plt.show()
How to Use Gaussian Naive Bayes in Scikit-Learn for Normal Distribution
Normal distribution has a mathematical equation that defines the probability of one observation of being in one of the groups. The formula is:
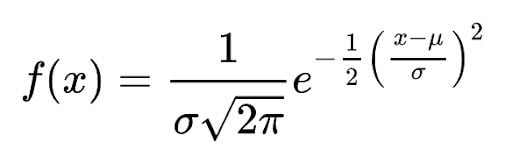
Where μ is mean and σ is standard deviation.
We can create a function to compute this probability:
#Creating a Function:
def normal_dist(x , mean , sd):
prob_density = (1/sd*np.sqrt(2*np.pi)) * np.exp(-0.5*((x-mean)/sd)**2)
return prob_density
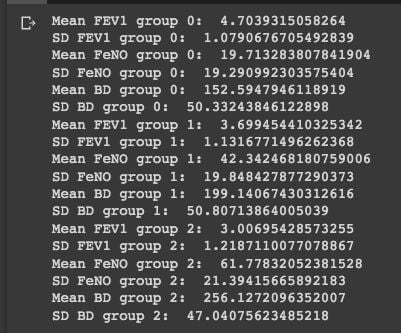
If we know the normal distribution formula, we can manually compute the probability of observation to be in one of the three groups. First, we need to compute the mean and standard deviation for all predictive parameters and groups:
#Group 0:
group_0 = df[df['dx'] == 0]
print('Mean FEV1 group 0: ', statistics.mean(group_0['FEV1']))
print('SD FEV1 group 0: ', statistics.stdev(group_0['FEV1']))
print('Mean FeNO group 0: ', statistics.mean(group_0['FeNO']))
print('SD FeNO group 0: ', statistics.stdev(group_0['FeNO']))
print('Mean BD group 0: ', statistics.mean(group_0['BD']))
print('SD BD group 0: ', statistics.stdev(group_0['BD']))
#Group 1:
group_1 = df[df['dx'] == 1]
print('Mean FEV1 group 1: ', statistics.mean(group_1['FEV1']))
print('SD FEV1 group 1: ', statistics.stdev(group_1['FEV1']))
print('Mean FeNO group 1: ', statistics.mean(group_1['FeNO']))
print('SD FeNO group 1: ', statistics.stdev(group_1['FeNO']))
print('Mean BD group 1: ', statistics.mean(group_1['BD']))
print('SD BD group 1: ', statistics.stdev(group_1['BD']))
#Group 2:
group_2 = df[df['dx'] == 2]
print('Mean FEV1 group 2: ', statistics.mean(group_2['FEV1']))
print('SD FEV1 group 2: ', statistics.stdev(group_2['FEV1']))
print('Mean FeNO group 2: ', statistics.mean(group_2['FeNO']))
print('SD FeNO group 2: ', statistics.stdev(group_2['FeNO']))
print('Mean BD group 2: ', statistics.mean(group_2['BD']))
print('SD BD group 2: ', statistics.stdev(group_2['BD']))
Now, using an example observation with:
FEV1 = 2.75FeNO = 27BD = 125
#Probability for:
#FEV1 = 2.75
#FeNO = 27
#BD = 125
#We have the same number of observations, so the general probability is: 0.33
Prob_geral = round(0.333, 3)
#Prob FEV1:
Prob_FEV1_0 = round(normal_dist(2.75, 4.70, 1.08), 10)
print('Prob FEV1 0: ', Prob_FEV1_0)
Prob_FEV1_1 = round(normal_dist(2.75, 3.70, 1.13), 10)
print('Prob FEV1 1: ', Prob_FEV1_1)
Prob_FEV1_2 = round(normal_dist(2.75, 3.01, 1.22), 10)
print('Prob FEV1 2: ', Prob_FEV1_2)
#Prob FeNO:
Prob_FeNO_0 = round(normal_dist(27, 19.71, 19.29), 10)
print('Prob FeNO 0: ', Prob_FeNO_0)
Prob_FeNO_1 = round(normal_dist(27, 42.34, 19.85), 10)
print('Prob FeNO 1: ', Prob_FeNO_1)
Prob_FeNO_2 = round(normal_dist(27, 61.78, 21.39), 10)
print('Prob FeNO 2: ', Prob_FeNO_2)
#Prob BD:
Prob_BD_0 = round(normal_dist(125, 152.59, 50.33), 10)
print('Prob BD 0: ', Prob_BD_0)
Prob_BD_1 = round(normal_dist(125, 199.14, 50.81), 10)
print('Prob BD 1: ', Prob_BD_1)
Prob_BD_2 = round(normal_dist(125, 256.13, 47.04), 10)
print('Prob BD 2: ', Prob_BD_2)
#Compute probability:
Prob_group_0 = Prob_geral*Prob_FEV1_0*Prob_FeNO_0*Prob_BD_0
print('Prob group 0: ', Prob_group_0)
Prob_group_1 = Prob_geral*Prob_FEV1_1*Prob_FeNO_1*Prob_BD_1
print('Prob group 1: ', Prob_group_1)
Prob_group_2 = Prob_geral*Prob_FEV1_2*Prob_FeNO_2*Prob_BD_2
print('Prob group 2: ', Prob_group_2)
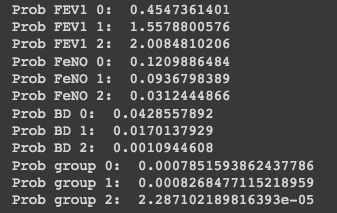
The results prove that our observation with the values FEV1 = 2.75, FeNO = 27, and BD = 125 has a higher probability of being in group two. While these steps are highly educational and great for learning, they are time-consuming.
Using Gaussian Naive Bayes in Scikit-Learn to Evaluate Normal Distribution
Fortunately, we have a much faster way to do it. We can use the Gaussian Naive Bayes from Scikit-Learn, which is similar to other classification algorithms in its implementation. We create X and y variables and perform train and test split:
#Creating X and y:
X = df.drop('dx', axis=1)
y = df['dx']
#Data split into train and test:
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.30)We also need to transform or normalize our data with standard scalar function:
sc = StandardScaler()
X_train = sc.fit_transform(X_train)
X_test = sc.transform(X_test)
And now build and evaluate the model:
#Build the model:
classifier = GaussianNB()
classifier.fit(X_train, y_train)
#Evaluate the model:
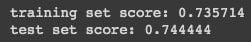
print("training set score: %f" % classifier.score(X_train, y_train))
print("test set score: %f" % classifier.score(X_test, y_test))
We can visualize our results using a confusion matrix:
# Predicting the Test set results
y_pred = classifier.predict(X_test)
#Confusion Matrix:
cm = confusion_matrix(y_test, y_pred)
print(cm)
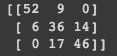
We can see by the confusion matrix that our model is best to predict class 0, but has a higher error rate with classes 1 and 2. Lastly, we can build decision boundaries graphs using two variables:
df.to_csv('data.csv', index = False)
data = pd.read_csv('data.csv')
def gaussian_nb_a(data):
x = data[['BD','FeNO',]].values
y = data['dx'].astype(int).values
Gauss_nb = GaussianNB()
Gauss_nb.fit(x,y)
print(Gauss_nb.score(x,y))
#Plot decision region:
plot_decision_regions(x,y, clf=Gauss_nb, legend=1)
#Adding axes annotations:
plt.xlabel('X_train')
plt.ylabel('y_train')
plt.title('Gaussian Naive Bayes')
plt.show()
def gaussian_nb_b(data):
x = data[['BD','FEV1',]].values
y = data['dx'].astype(int).values
Gauss_nb = GaussianNB()
Gauss_nb.fit(x,y)
print(Gauss_nb.score(x,y))
#Plot decision region:
plot_decision_regions(x,y, clf=Gauss_nb, legend=1)
#Adding axes annotations:
plt.xlabel('X_train')
plt.ylabel('y_train')
plt.title('Gaussian Naive Bayes')
plt.show()
def gaussian_nb_c(data):
x = data[['FEV1','FeNO',]].values
y = data['dx'].astype(int).values
Gauss_nb = GaussianNB()
Gauss_nb.fit(x,y)
print(Gauss_nb.score(x,y))
#Plot decision region:
plot_decision_regions(x,y, clf=Gauss_nb, legend=1)
#Adding axes annotations:
plt.xlabel('X_train')
plt.ylabel('y_train')
plt.title('Gaussian Naive Bayes')
plt.show()
gaussian_nb_a(data)
gaussian_nb_b(data)
gaussian_nb_c(data)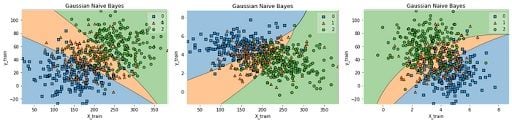
And that’s all there is to it!
Frequently Asked Questions
When to use Gaussian Naive Bayes?
The Gaussian Naive Bayes classifier works best for situations involving continuous data that is arranged as a normal distribution, meaning the data points are set up symmetrically to form a bell curve shape.
What is the difference between Gaussian Naive Bayes and Multinomial Naive Bayes?
The main difference between Gaussian Naive Bayes and Multinomial Naive Bayes is the types of data they work with. Gaussian Naive Bayes works with continuous data — data that forms a constant sequence, meaning it can take any value within a range. Multinomial Naive Bayes works with discrete data — data that has spaces between values. Discrete data is counted rather than measured, while continuous data is measured but not countable.
Does Naive Bayes assume Gaussian distribution?
In general, Naive Bayes classifiers do not assume that data follows a Gaussian, or normal, distribution. Gaussian Naive Bayes is the only Naive Bayes classifier that assumes a Gaussian distribution.
How do you know if a distribution is Gaussian?
If the mean, median and mode are equal, then the data fits a Gaussian distribution. You can also plot the data points, and they should form the shape of a symmetrical bell curve.





