One hot encoding (OHE) is a machine learning technique that encodes categorical data to numerical ones. If you want to perform one hot encoding, both sklearn.preprocessing.OneHotEncoder and pandas.get_dummies are popular choices.
One Hot Encoding Definition
One hot encoding is a machine learning technique that encodes categorical data into numerical ones. It’s used to give weight to categorical data so that it can be used in a linear regression model.
Most data scientists recommend using Scikit-learn (sklearn) because its fit/transform paradigm provides a built-in mechanism to learn all the possible categories from the training set and apply them to the validation or real input data. Therefore, this approach will prevent errors arising when the validation or real input data does not contain all categories, or the categories do not appear in the same order.
In this article I will argue that there is no clear winner of this competition. For data scientists who use Pandas DataFrame, using the native Pandas get_dummies function has clear benefits, and there is a very straightforward way to avoid the above mentioned issue.
What Is One Hot Encoding?
One hot encoding (OHE) is a technique that encodes categorical data to numerical ones. It’s mainly used in machine learning applications. If, you’re building a model to predict the weight of animals, one of your inputs is going to be the type of animal, i.e. cat, dog or parrot. This is a string value, and therefore, models like linear regression aren’t able to deal with it.
The approach that first comes to mind is to give integer labels to the animals and replace each string with the corresponding integer representation. But if you do this, you introduce an artificial ordering of the animals. For example, a parrot will have three times more impact for the “animal” weight than a cat. Instead, OHE creates a new input variable, such as a column, for each of the animals and sets this variable to one or zero depending on whether the animal is the selected one. For example:
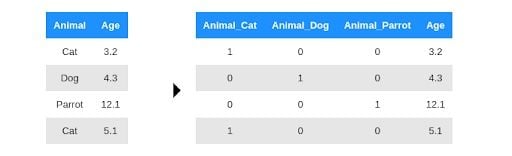
After this separation, your linear model can assign weights to these new columns independently of the others. In practice, you don’t really need three columns to represent the three animals. You can choose any one of them to drop. In other words, if it’s not a dog or a cat, it must be a parrot.
One Hot Encoding: Scikit vs Pandas
Both sklearn and Pandas provide methods to perform this operation, and there’s a debate with a long history among data scientists about which one to use. The reason I want to revisit the topic is because both of these libraries have evolved, and there are new features that are worth taking into account when deciding which one to use for OHE.
There are several options one may specify when encoding, such as whether to use sparse or dense data representation or whether to keep all new columns or drop one of them. Both libraries support many of such features, but I won’t focus on those. My main focus will be how each handles the categories below.
If you do a train/test split, either manually or automated using sklearn.model_selection.train_test_split, it may easily happen that your train data set won’t contain any parrots. It’s not theoretically an issue If some categories are missing. You can still make a prediction, though, probably less accurately. But your code will break if it’s not prepared for this difference, as the columns in the fitted data don’t agree with the columns of the data used for prediction.
In this article, I’ll focus on the following points:
- How do you tell the OHE the set of all categories, and how do you make sure the encoding is applied consistently to train, test and validate real data?
- How do you apply the encoding to a Pandas DataFrame?
- How do you incorporate the one hot encoding in a sklearn pipeline?
One Hot Encoding Using Scikit-Learn
The usual wisdom is to use sklearn’s sklearn.preprocessing.OneHotEncoder for this purpose, because using its fit/transform paradigm allows you to use the training data set to “teach” categories and apply it to your real-world input data.
The main steps are the followings:
enc = OneHotEncoder(...)
encoded_X_train= enc.fit_transform(X_train['Animal'])
encoded_input = enc.transform(real_input['Animal'])
Where X_train is your training input data and real_input is your real input data to which you want to apply the model.
If you are lucky, then all possible categories will appear in X_train. The encoder object learns these categories and the corresponding mappings, and it will produce the right columns in the right column order for the real input. However, sklearn.preprocessing.OneHotEncoder does produce a NumPy array, so the order of the columns is important.
But you shouldn’t assume that you’ll always be lucky. For example, if you use cross-validation to randomly and repeatedly split your data into train and test parts, you may easily end up in a situation where your actual training data is missing some of the categories. This leads to errors, as you won’t be able to transform the data in the test set.
Sklearn’s solution for this is to explicitly provide the possible categories to the OneHotEncoder object as follows:
enc = OneHotEncoder(..., categories=[['cat'/'dog'/'parrots']]).
You need to provide a list of lists in the categories parameter in order to specify the categories for each of the input columns.
Another common step, when using sklearn is to do the conversion between raw NumPy arrays and Pandas DataFrames. You can either use sklearn.compose.make_column_transformer for this, or implement it manually, using the .get_feature_names_out() method of OneHotEncoder to give you the column names for the new features. Let’s see examples for both of these. I will add another column, Color, in order to make the examples more informative.
import pandas as pd
from sklearn.preprocessing import OneHotEncoder
from sklearn.compose import make_column_transformer
from sklearn.pipeline import make_pipeline
Specifying Inputs and Encoder
# It is always good to have an explicit list of categories
categories = [('Type',['Cat','Dog','Parrot','Whale']),
('Color',['Brown','Black','Mixed'])]
# Let's create some mock data
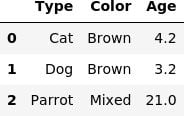
X_train = pd.DataFrame(columns=['Type','Color','Age'],
data=[['Cat','Brown',4.2],['Dog','Brown',3.2],['Parrot','Mixed',21]])
X_input = pd.DataFrame(columns=['Type','Color','Age'],
data = [['Parrot', 'Black', 32]])
display(X_train, X_input)

ohe_columns = [x[0] for x in categories]
ohe_categories = [x[1] for x in categories]
enc = OneHotEncoder(sparse_output=False, categories=ohe_categories)
Column Transformer Approach
# We create a column transformer telling it to replace the columns which hold the categories and leave the rest untouched.
# The column transformer does not create the pandas DataFrame, but it selects the appropriate columns, converts them and appends the converted columns to the other ones.
transformer = make_column_transformer((enc, ohe_columns), remainder='passthrough')
# We convert the resulting arrays to DataFrames
transformed=transformer.fit_transform(X_train)
display(pd.DataFrame(
transformed,
columns=transformer.get_feature_names_out(),
index=X_train.index
))
pd.DataFrame(
transformer.transform(X_input),
columns=transformer.get_feature_names_out(),
index=X_input.index
)
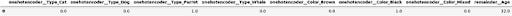
We can see that the column converter does part of the job, but we still need to do additional work if we want to use DataFrames. Also, I don’t really like these column names, but there’s no way to tune them, other than manual post processing. Note, that columns are created for all possible categories, not only those that appear in the input.
Manual Approach
I call it manual, as we use the OneHotEncoder object directly and deal with selecting, and appending the columns ourselves.
transformed_df = pd.DataFrame(
enc.fit_transform(X_train[ohe_columns]),
columns = enc.get_feature_names_out(),
index = X_train.index)
transformed_df = pd.concat([X_train.drop(ohe_columns,axis=1),transformed_df],axis=1)
display(transformed_df)
T_input = pd.DataFrame(
enc.transform(X_input[ohe_columns]),
columns = enc.get_feature_names_out(),
index = X_input.index)
T_input = pd.concat([X_input.drop(ohe_columns,axis=1),T_input],axis=1)
T_input
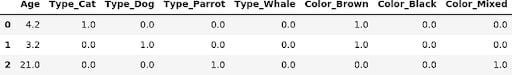

We had to do a bit more manual work, but the column names are much more friendly. In newer versions of sklearn (1.3 and above), we can fine-tune these names.
Pipelines
A scikit pipeline is a convenient way to sequentially apply a list of transforms. You can use it to assemble several steps that can be cross-validated together while setting different parameters.
The manual/raw approach is generally not suited to be included in a pipeline because of the additional steps needed to select and add the columns. The column transformer approach, on the other hand, is suited for pipelines. The additional steps we made were only required to transform the NumPy array to DataFrame, which is not a requirement for a pipeline.
One Hot Encoding in Pandas
The pandas.get_dummies function does not follow the fit/transform model, nor does it have an explicit input parameter specifying the available categories. One could conclude that it’s inappropriate for the job. This conclusion, however, is not correct.
Pandas inherently supports the handling of categorical data through pandas.CategoricalDtype. You need to do your homework and set up the column categories properly. Once that is done consistently, you no longer need the fitting step.
Using the categorical type has additional benefits, like reduced storage space and checking for typos. Let’s see how this is done:
X_train[ohe_columns] = X_train[ohe_columns].astype('category')
X_input[ohe_columns] = X_input[ohe_columns].astype('category')
for column_name, l in categories:
X_train[column_name] = X_train[column_name].cat.set_categories(l)
X_input[column_name] = X_input[column_name].cat.set_categories(l)
Now, all we need to do is to call the get_dummies function.
display(pd.get_dummies(X_train))
display(pd.get_dummies(X_input))
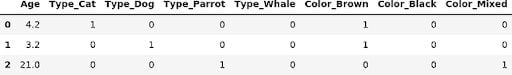

As we can see, after the categories are properly set, there is no additional work needed to have a nice DataFrame. Actually, I did a bit of cheating above: by default, get_dummies converts all columns with object, string or category dtype. If this isn’t what we want, we can explicitly specify the list of columns to convert using the columns parameter of get_dummies:
pd.get_dummies(X_train, columns=ohe_columns[:1])
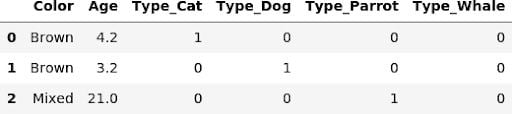
In order for a transformer to be eligible for a pipeline it has to implement the fit and transform methods, which the get_dummies function clearly doesn’t do. Fortunately, it’s super easy to create a custom transformer for this task:
Now, we can use our new class as any other sklearn transformer, we can even embed it in a pipeline.
from sklearn.base import BaseEstimator, TransformerMixin
class GetDummiesTransformer(BaseEstimator, TransformerMixin):
def __init__(self, *args, pandas_params={}, **kwargs):
super().__init__(*args, **kwargs)
self._pandas_params = pandas_params
def fit(self, X, y=None):
return self
def transform(self, X, y=None):
return pd.get_dummies(X, **self._pandas_params)
When writing this transformer we assumed that the relevant columns already have categorical dtypes. But it’s very simple to add a few lines of code to GetDummiesTransformer to allow the specification of the columns in the __init__ function.
Advantages and Disadvantages to One Hot Encoding in Scikit-Learn and Pandas
As we have seen it is possible and very much suggested to explicitly specify the available categories for both the scikit OneHotEncoder and the pandas get_dummies approaches. Remember: Explicit is better than implicit. This means that both of these approaches are well suited for the task, so it’s a personal preference. For sklearn, the explicit category setting was achieved by passing a parameter to the constructor of the OneHotEncoder class, while for Pandas, we had to set up the categorical data type.
- Using the “raw” version of
OneHotEncoder, i.e. without a column transformer, needs the most manual adjustment, and I only see rare cases in which I would use this approach in practice. - If your process relies on scikit pipelines, which has many advantages, then using scikit
OneHotEncoderwith a column transformer seems to be the most natural choice to me. - If you like to process the data step-by-step, going from DataFrame to DataFrame, which can be a good choice in the exploration phase, then I would definitely take the
pandas.get_dummiesapproach.





