Over the past decade, the adoption of electronic health record (EHR) systems in hospitals has become widespread. This transformation is due to the Health Information Technology for Economic and Clinical Health (HITECH) Act of 2009, which allocated $30 million in incentives for hospitals and physician practices to adopt EHR systems. This digital explosion in big healthcare data lends itself to modern machine learning tools that can be used for a variety of tasks such as disease detection, patient journey tracking, concept representation, patient de-identification, and data augmentation. In the past, a class of deep learning models, called convolutional neural networks (CNNs) have been successfully deployed toward a variety of disease detection tasks, including classification tasks related to interstitial lung disease and colonoscopy frames and detection of polyps and pulmonary embolisms.
These efforts have been successful because the underlying data for these tasks typically contain sufficient positive and negative examples for each detection class. In each of the example diseases/disorders above, lots of people test positive and lots also test negative. Having numerous positive and negative examples helps the machine model to learn more effectively. For disease detection problems with an imbalance in positive versus negative results, supervised learning methods like CNNs struggle to perform. For example, supervised machine learning models might struggle with a rare disease like Ebola because very few patients will test positive for it, leading to a much larger group of negatives.
Generative Adversarial Networks (GANs) are useful in these cases because they can learn to produce fake examples of the underrepresented data, better training the model. In addition to improving disease detection, GANs can be used for data de-identification, which prevents patient personal information from exposure.The Health Insurance Portability and Accountability Act of 1996 (HIPAA) Privacy Rule mandates protection of patient information, meaning that healthcare providers need to take it seriously. Data de-identification is a challenging problem in the healthcare analytics space because traditional methods are not robust enough to withstand re-identification. Namely, most current methods of de-identification can be reversed, compromising the privacy of healthcare patients’ personal records. GAN models, both in research and practice, suggest promising solutions to many of the thorny problems facing healthcare today.
What Are Generative Adversarial Networks?
Before we get into the healthcare applications of GANs, let’s discuss some of the basics of their operation. The most important underlying concepts behind GANs are deep neural networks (namely convolutional neural networks) and backpropagation. Given their relevance, we should briefly review these terms.
Artificial neural networks are mathematical models loosely inspired by the human brain. They are composed of a collection of units (artificial neurons) and connections between units (synapses), which each have weights that vary as learning improves through the process of backpropagation. To understand backpropagation, consider the illustration below. The error values associated with a neural network’s predictions are plotted against the weight values assigned to “learned” features. The goal of training a neural network is to find the weights that give the smallest error, which corresponds to finding the bottom of the hill in the figure. You can picture the learning process to be similar to walking around the hilly terrain until you find the lowest point.
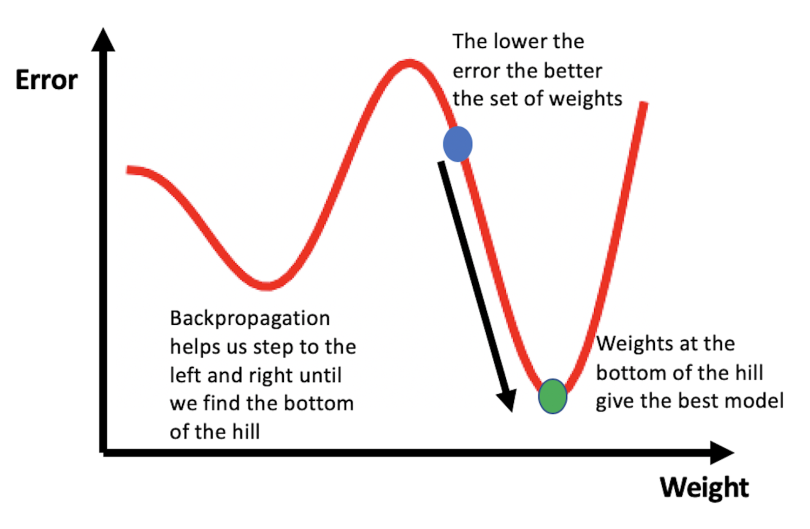
A typical neural network is a collection of these interconnected and weighted units, which allow the models to capture high dimensional and nonlinear relationships between input and output. This structure is part of the reason why they work so well. In the context of image analysis, low-level features generated by a neural network may be edges (i.e., the edges or outline of a face) and high-level features may be concepts identifiable by humans, such as letters, digits or whole faces.
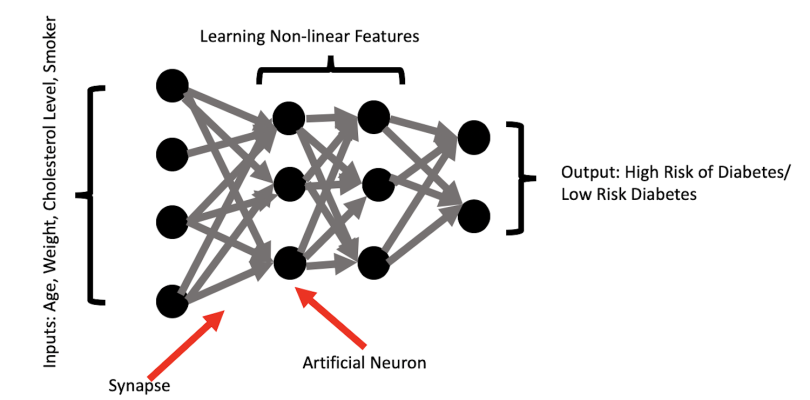
The illustration below shows a simple example of predicting risk for diabetes based on patient characteristics such as age, weight, cholesterol level, and smoker status. The neural network performs transformations on these inputs and engineers important predictive features that are nonlinear combinations of the inputs. The network then uses the learned features to predict patient risk for diabetes.
An example of a high-level feature in this context may be capturing the compounding effect of being an overweight senior who is a heavy smoker with high cholesterol. The diabetes risk of this patient is probably significantly higher than a young, overweight heavy smoker with high cholesterol. But what about an obese senior who is a non-smoker with high cholesterol? Is the obese non-smoker more or less at risk than the overweight heavy smoker? A human would have a hard time accurately predicting the outcome for these two patients. Fortunately, neural networks excel at capturing the complex interactions between these features and how they affect the probability of an outcome.
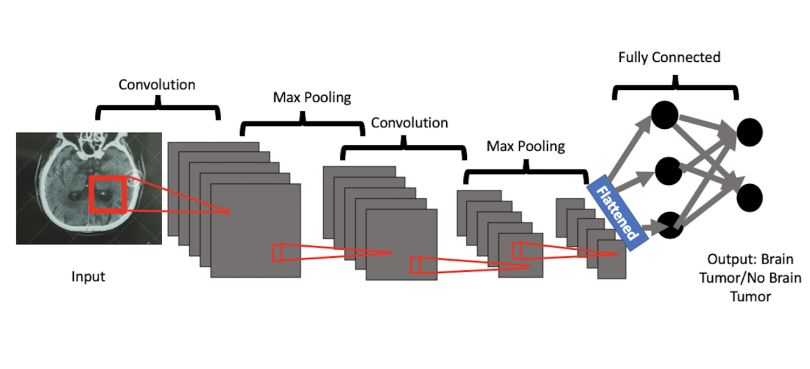
Convolutional neural networks are extensions of these simpler networks, but they also include convolutional layers and pooling layers. At a high level, these layers excel at learning the most important parts of images as they relate to the content of the overall image. As I alluded to earlier, these high-level features may be things that humans could identify. The convolution layer extracts the high-level concepts like faces, letters, objects, or digits, and the max pooling layer reduces those features to the most essential ones required for learning.
The image below shows the example of classifying whether or not a brain tumor is present in a patient’s CT scan. The convolutional neural network uses the convolution layer to learn high-level features from the image that most closely map to both healthy brains and brains with tumors. Relevant features may include segments of healthy brain tissue, segments of tumor, the whole tumor, the edges of the tumor, and additional features that may not necessarily be interpretable by humans but are useful for machine learning. The max pooling layers reduce these extracted features by removing redundant image segments that don’t improve learning. This process helps the model prioritize the most essential or most commonly present patches of images, represented by high-level features, from the image pixels.
Now that we have the basics, let’s discuss GANs. GANs are a set of deep neural network models, developed by Ian Goodfellow in 2014, used to produce synthetic data. Synthetic data is any data that is relevant to a specific situation that was not directly obtained through real-world measurements.
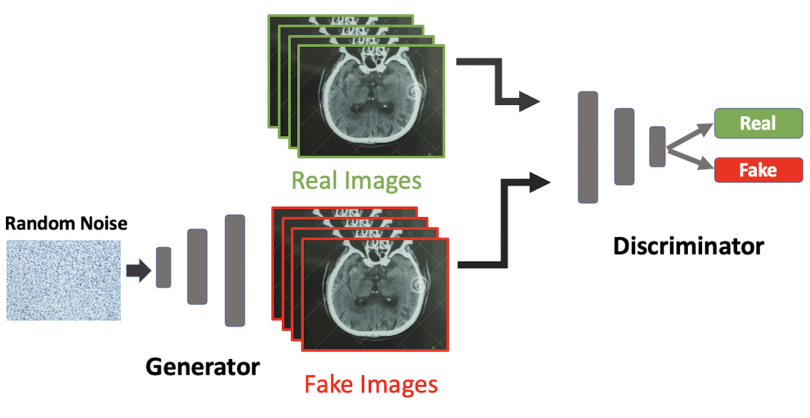
The goal of a GAN is to train a discriminator to be able to distinguish between real and fake data while simultaneously training a generator to produce synthetic instances of data that can reliably trick the discriminator. The discriminator is an ordinary convolutional neural network used to distinguish between authentic and synthetic images, and the generator is a modified convolutional neural network that is trained to produce authentic-looking fake images. GANs train the discriminator and generator together to iteratively improve the ability of the discriminator to spot fake images and the ability of the generator to produce realistic images.
In the illustration below, a generator is trained on random noise pixel data and then generates fake brain CT images. Fake images made by the generator are then fed into the discriminator alongside real images. The discriminator is an ordinary CNN that’s been trained to distinguish between real and fake images. The generator’s training aims to produce increasingly realistic fake images that can trick the discriminator. As training continues, the generator gets better at producing fakes, and the discriminator gets better at distinguishing between real and fake images until the generator produces images that closely resemble the authentic images. Once training is complete, the GAN should be able to produce realistic images that can be used to augment existing data or create entirely new data sets. Data augmentation is useful for situations where there is an imbalance in examples (e.g., data corresponding to rare diseases like Ebola). Further, creating new synthetic data sets is useful for protecting patient privacy as well providing an alternative to purchasing expensive clinically annotated medical data.
GANs in Medical Imaging
The most prevalent applications of GANs in healthcare involve medical imaging. Two important medical imaging tasks are data segmentation for brain tumors and medical image synthesis.
Brain Tumor Segmentation
An interesting application of GANs is toward brain tumor segmentation, which amounts to partitioning brain CT images into image objects like tumor edges, healthy tissue, and whole tumor sites. Although detecting most types of brain tumor, such as low- and high-grade gliomas, is typically straightforward for doctors, defining the tumor border by visual assessment remains a challenge. In a recent study, Eklund et. al. developed a method called Vox2Vox in an effort to perform brain tumor segmentation. In their study, the researchers trained an ensemble of 3D GAN models on the Brain Tumor Segmentation (BraTS) Challenge 2020 data set and were able to generate high-quality brain tumor segments. Specifically, their ensemble model was able to detect whole tumors, tumor cores, and active tumors with dice coefficients > 87 percent for each class, which outperforms past studies that use CNNs. These methods provide means for generating quality brain tumor segments that can be helpful for doctors during analysis, treatment and surgery.
Medical Image Synthesis
Another interesting application of GANs is in medical image synthesis. Medical imaging data is rarely available for large-scale analysis due to the high cost of obtaining clinical annotations. Given this blocker, many research projects have worked to develop reliable methods for medical image synthesis. In a 2017 paper, Campilho et. al. developed a method for generating synthetic retinal images. By training a GAN on retinal images from the DRIVE database, these researchers were able to demonstrate the feasibility of generating quality retinal images that don’t truly exist. These fake images can be used to augment data in situations where the number of retinal images is limited and can further be used to train future AI models. In the future, these methods can be used to produce data for training models to detect diseases where there isn’t enough real data to train an accurate model. Additionally, this type of synthetic imaging data can be used to further protect patient privacy.
Generating Synthetic Discrete Medical Data
While the most commonplace use of GANs is toward medical image synthesis and image segmentation, GANs can also help with generating synthetic multi-label discrete data. This is a useful healthcare application of GANs as it can provide robust protection of patient privacy. Typically, in the effort to protect patient privacy, methods for de-identification of EHR records include slightly modifying personal identifiable attributes such as data of birth. A common method of de-identification is generalization, in which an attribute like data of birth can be generalized to only the month and year.
Unfortunately, in this method, a one-to-one mapping still exists between the altered records and the originals from which they were derived. That makes them vulnerable to attacks where bad actors can re-identify patient information. An alternative approach involves developing synthetic patient data. In 2017, Sun et. al. developed medical GANs (medGAN), to generate synthetic multi-label discrete patient records. Using this method, they were able to accurately produce synthetic binary and count variables that represent events in EHRs like the diagnosis of a certain disease or treatment of a certain medication. Given how difficult it is to access EHR data, medGAN is a significant contribution to healthcare research as it provides a way to generate quality synthetic patient data. The authors also provided an empirical evaluation of privacy and demonstrated that medGAN posed limited risk in personal identification, which further validates their method.
Conclusions
Given that GANs have shown promise in the space of image segmentation, image synthesis and discrete patient data synthesis, they have the potential to revolutionize healthcare analytics. The method of image segmentation in medicine can be extended to foreign object identification in medical images, detection of additional types of tumor growths, and precise identification of organ structure. Toward the latter, image segmentation using GANs can be used to give precise structural details of the brain, liver, chest, abdomen and liver in MRIs. Further, GANs show strong promise in the space of medical image synthesis. In many instances, medical imaging analysis is limited by lack of data and/or high cost of authentic data. GANs can circumvent these issues by enabling researchers and doctors to work with high-quality, realistic synthetic images. This can significantly improve disease diagnosis, prognosis and analysis. Finally, GANs show significant potential in the space of patient data privacy, as it provides a more reliable way to implicitly map real patient data to synthetic data. This implicit map improves the data privacy of patients since it is not a typical one-to-one map, and is thereby difficult to recover explicitly. Improvements in medical image segmentation, image synthesis and data anonymization are all stepping stones toward improving the efficiency and reliability of healthcare informatics.





