Finally, my program is running! Should I go and get a coffee?
Data scientists have laptops with quad-core or octa-core processors and Turbo Boost technology. We routinely work with servers with even more cores and computing power. But do we really use the raw power we have at hand?
Instead of taking advantage of our resources, too often we sit around and wait for time-consuming processes to finish. Sometimes we wait for hours, even when urgent deliverables are approaching the deadline. Can we do better?
In this post, I will explain how to use multiprocessing and Joblib to make your code parallel. Using these libraries, we will be able to use the multiple cores of our machines, making our code run faster.
Parallel Processing in Data Science
Using Multiprocessing With a Single Parameter Function
We’ll start with a problem where we have a big list of items, and we want to apply a function to every element on the list.
Why do we want to do this? Although it may seem like a trivial problem, this is what we do on a daily basis in data science. For example, we may have a model, and we want to run multiple iterations of the model with different hyperparameters. Or we’re creating a new feature in a big data frame, and we need to apply a function row by row to the data frame using the apply keyword. By the end of this post, you will be able to parallelize most of the use cases you face in data science with this simple construct.
So, coming back to our hypothetical problem, let’s say we want to apply the square function to all our elements in the list.
def square(num):
return x**2Of course, we can use simple Python to run this function on all elements of the list.
result = [f(x) for x in list(range(100000))]But the code is running sequentially. That means that only one core in our machine is doing all the work. Theoretically, we can share this load with all the cores in our machine. To do so, we can use multiprocessing to apply this function to all the elements of a given list in parallel using the eight cores in our powerful computer.
from multiprocessing import Pool
pool = Pool(8)
result = pool.map(f,list(range(100000)))
pool.close()These lines create a multiprocessing pool of eight workers, and we can use this pool to map our required function to this list.
Let’s check to see how this code performs:
from multiprocessing import Pool
import time
import plotly.express as px
import plotly
import pandas as pd
def f(x):
return x**2
def runner(list_length):
print(f"Size of List:{list_length}")
t0 = time.time()
result1 = [f(x) for x in list(range(list_length))]
t1 = time.time()
print(f"Without multiprocessing we ran the function in {t1 - t0:0.4f} seconds")
time_without_multiprocessing = t1-t0
t0 = time.time()
pool = Pool(8)
result2 = pool.map(f,list(range(list_length)))
pool.close()
t1 = time.time()
print(f"With multiprocessing we ran the function in {t1 - t0:0.4f} seconds")
time_with_multiprocessing = t1-t0
return time_without_multiprocessing, time_with_multiprocessing
if __name__ == '__main__':
times_taken = []
for i in range(1,9):
list_length = 10**i
time_without_multiprocessing, time_with_multiprocessing = runner(list_length)
times_taken.append([list_length, 'No Mutiproc', time_without_multiprocessing])
times_taken.append([list_length, 'Multiproc', time_with_multiprocessing])
timedf = pd.DataFrame(times_taken,columns = ['list_length', 'type','time_taken'])
fig = px.line(timedf,x = 'list_length',y='time_taken',color='type',log_x=True)
plotly.offline.plot(fig, filename='comparison_bw_multiproc.html')
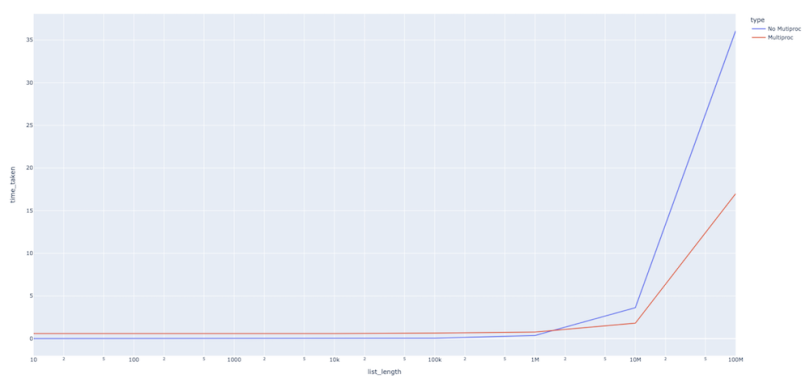
As we can see, the multiprocessing runtime was somewhat higher for some list length, but it doesn’t increase as fast as the non-multiprocessing function runtime increases for larger list lengths. This tells us that using multiprocessing comes with a certain amount of computational overhead, so using it for processes that don’t take much time doesn’t make a whole lot of sense.
In practice, we won’t use multiprocessing for functions that conclude in milliseconds, but, rather, for much larger computations that could take much more than a few seconds, sometimes up to hours.
So, let’s try a more involved computation, which would normally take more than two seconds. I am using time.sleep as a proxy for computation here.
from multiprocessing import Pool
import time
import plotly.express as px
import plotly
import pandas as pd
def f(x):
time.sleep(2)
return x**2
def runner(list_length):
print(f"Size of List:{list_length}")
t0 = time.time()
result1 = [f(x) for x in list(range(list_length))]
t1 = time.time()
print(f"Without multiprocessing we ran the function in {t1 - t0:0.4f} seconds")
time_without_multiprocessing = t1-t0
t0 = time.time()
pool = Pool(8)
result2 = pool.map(f,list(range(list_length)))
pool.close()
t1 = time.time()
print(f"With multiprocessing we ran the function in {t1 - t0:0.4f} seconds")
time_with_multiprocessing = t1-t0
return time_without_multiprocessing, time_with_multiprocessing
if __name__ == '__main__':
times_taken = []
for i in range(1,10):
list_length = i
time_without_multiprocessing, time_with_multiprocessing = runner(list_length)
times_taken.append([list_length, 'No Mutiproc', time_without_multiprocessing])
times_taken.append([list_length, 'Multiproc', time_with_multiprocessing])
timedf = pd.DataFrame(times_taken,columns = ['list_length', 'type','time_taken'])
fig = px.line(timedf,x = 'list_length',y='time_taken',color='type')
plotly.offline.plot(fig, filename='comparison_bw_multiproc.html')
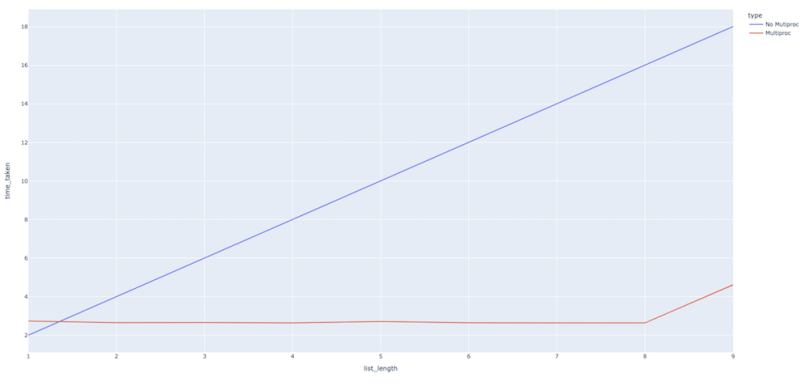
As we can see, the difference is much more stark in this case. The function takes much more time without multiprocessing than when we use it. Again, this makes perfect sense because, when we start multiprocessing, eight workers start working in parallel on the tasks instead of their happening in a sequential manner, with each task taking two seconds.
Multiprocessing With Multiple Params Function
An extension to the code above occurs in cases where we have to run a function that could take multiple parameters. For a use case, let’s say you have to tune a particular model using multiple hyperparameters. You can do something like this:
import random
def model_runner(n_estimators, max_depth):
# Some code that runs and fits our model here using the
# hyperparams in the argument.
# Proxy for this code with sleep.
time.sleep(random.choice([1,2,3])
# Return some model evaluation score
return random.choice([1,2,3])How would you run such a function? You can do this in two ways.
Using Pool.map and * magic
def multi_run_wrapper(args):
return model_runner(*args)
pool = Pool(4)
hyperparams = [[100,4],[150,5],[200,6],[300,4]]
results = pool.map(multi_run_wrapper,hyperparams)
pool.close()In this code, we provide args to the model_runner using
Using pool.starmap
From Python3.3 onwards, we can use the starmap method to accomplish this even more easily.
pool = Pool(4)
hyperparams = [[100,4],[150,5],[200,6],[300,4]]
results = pool.starmap(model_runner,hyperparams)
pool.close()
Using Joblib With a Single Parameter Function
Joblib is another library that provides a simple helper class to write parallel for loops using multiprocessing. I find it much easier to use than the multiprocessing module. Running a parallel process is as simple as writing a single line with the parallel and delayed keywords:
from joblib import Parallel, delayed
import time
def f(x):
time.sleep(2)
return x**2
results = Parallel(n_jobs=8)(delayed(f)(i) for i in range(10))Let’s compare Joblib parallel to the multiprocessing module using the same function we used before.
from multiprocessing import Pool
import time
import plotly.express as px
import plotly
import pandas as pd
from joblib import Parallel, delayed
def f(x):
time.sleep(2)
return x**2
def runner(list_length):
print(f"Size of List:{list_length}")
t0 = time.time()
result1 = Parallel(n_jobs=8)(delayed(f)(i) for i in range(list_length))
t1 = time.time()
print(f"With joblib we ran the function in {t1 - t0:0.4f} seconds")
time_without_multiprocessing = t1-t0
t0 = time.time()
pool = Pool(8)
result2 = pool.map(f,list(range(list_length)))
pool.close()
t1 = time.time()
print(f"With multiprocessing we ran the function in {t1 - t0:0.4f} seconds")
time_with_multiprocessing = t1-t0
return time_without_multiprocessing, time_with_multiprocessing
if __name__ == '__main__':
times_taken = []
for i in range(1,16):
list_length = i
time_without_multiprocessing, time_with_multiprocessing = runner(list_length)
times_taken.append([list_length, 'No Mutiproc', time_without_multiprocessing])
times_taken.append([list_length, 'Multiproc', time_with_multiprocessing])
timedf = pd.DataFrame(times_taken,columns = ['list_length', 'type','time_taken'])
fig = px.line(timedf,x = 'list_length',y='time_taken',color='type')
plotly.offline.plot(fig, filename='comparison_bw_multiproc.html')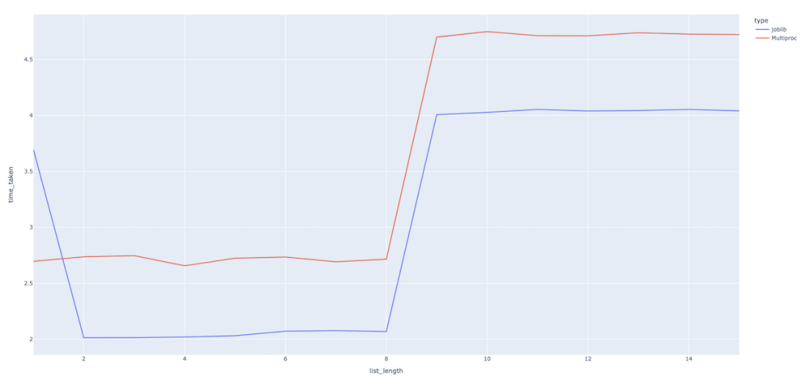
We can see that the runtimes are pretty much comparable. Even better, the Joblib code looks much more succinct than that of the multiprocessing approach.
Using Joblib With Multiple Params Function
Using multiple arguments for a function is as simple as just passing the arguments using Joblib. Here is a minimal example.
from joblib import Parallel, delayed
import time
def f(x,y):
time.sleep(2)
return x**2 + y**2
params = [[x,x] for x in range(10)]
results = Parallel(n_jobs=8)(delayed(f)(x,y) for x,y in params)
Save Time With Multiprocessing
Multiprocessing is a pretty nice concept and something that every data scientist should at least know about. It won’t solve all your problems, and you should still work on optimizing your functions. Still, having it in your toolkit will save a lot of time that you would otherwise spend just waiting for your code to finish or staring at that screen when you could have already taken the results and presented them to the business.
Also, if you want to learn more about Python 3, I would like to call out this excellent course from the University of Michigan. Do check it out.





